Exploring the Weather in Athens¶
In this assignment we explore the weather in Athens over a period of more than 50 years. This assignment was done as a project for the Practical Data Science course taught by Mr. Panos Louridas at the Athens University of Economics and Business.
Questions¶
Q1: Obtain the Data¶
We work with data covering the period from 1955 to 2020. The data are obtained from two sources:
-
Data downloaded from the National Oceanic and Atmospheric Administration's National Centers for Environmental Information (https://www.ncdc.noaa.gov/cdo-web/) and in particular https://www.ncdc.noaa.gov/cdo-web/search.
-
As we are focusing on Athens, we use the data from the Hellinikon weather station and we concentrate on the average daily temperature and precipitation.
-
We firstly explore the completeness of the data. What data are missing?
-
To fill in any missing data we will will use an alternative dataset available from https://data.hellenicdataservice.gr/dataset/66e1c19a-7b0e-456f-b465-b301a1130e3f; this dataset covers only the period from 2010-2019.
To download the data from the National Oceanic and Atmospheric Administration's National Centers for Environmental Information we follow the steps below.
- We start by navigating to the https://www.ncdc.noaa.gov/cdo-web/search and
inputting the required fields for our search as seen below.
Here we fill the fields with values:
- Dataset: Daily Summaries
- Date Range: 1955-01-01 to 2019-12-31
- Search for: Stations
- Search Term: Hellinikon
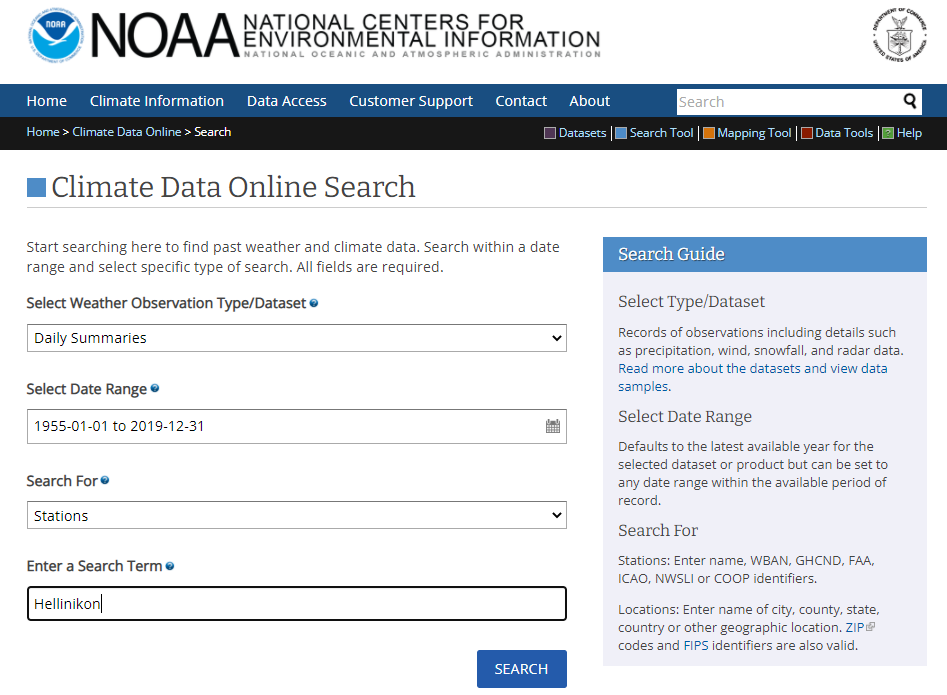
- In the page that loads, we select the Hellinikon station in the upper left side.
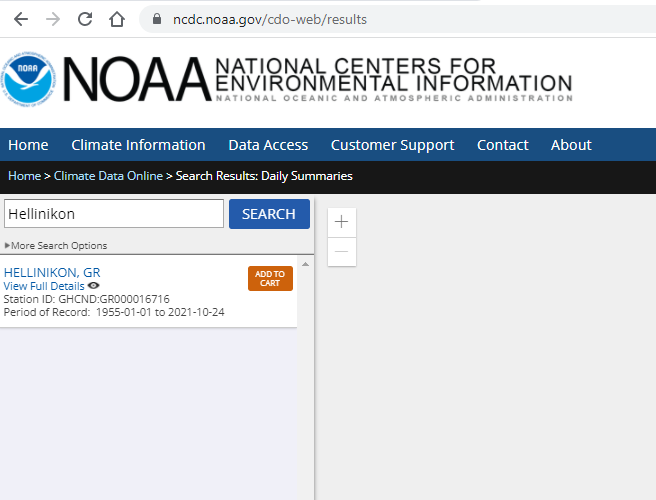
- Next we add the dataset to the cart
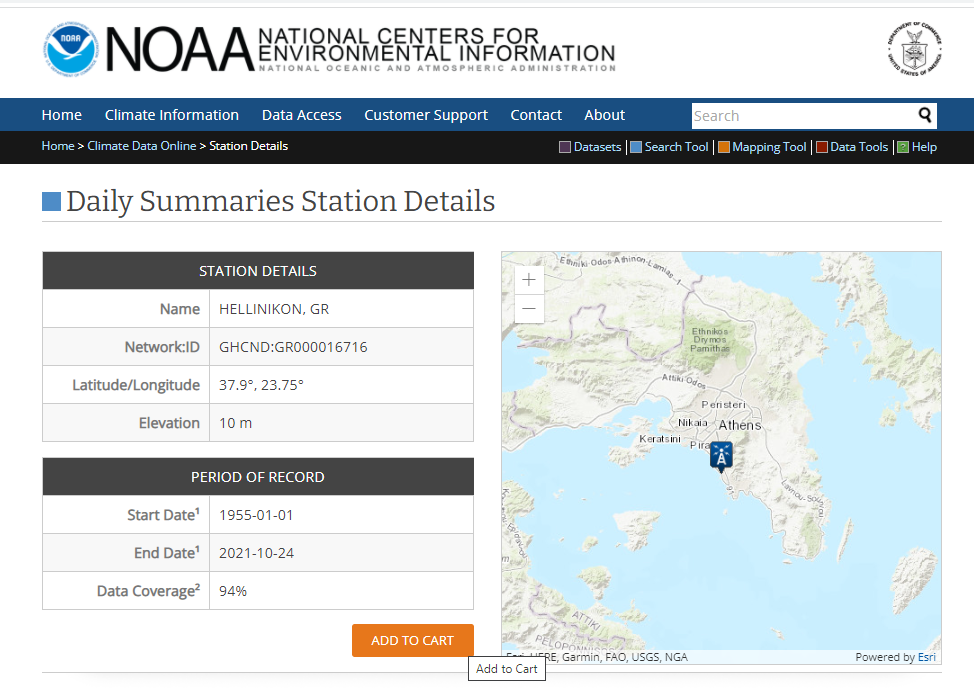
- In the cart options we select csv format and the date range for which we want to collect the
data
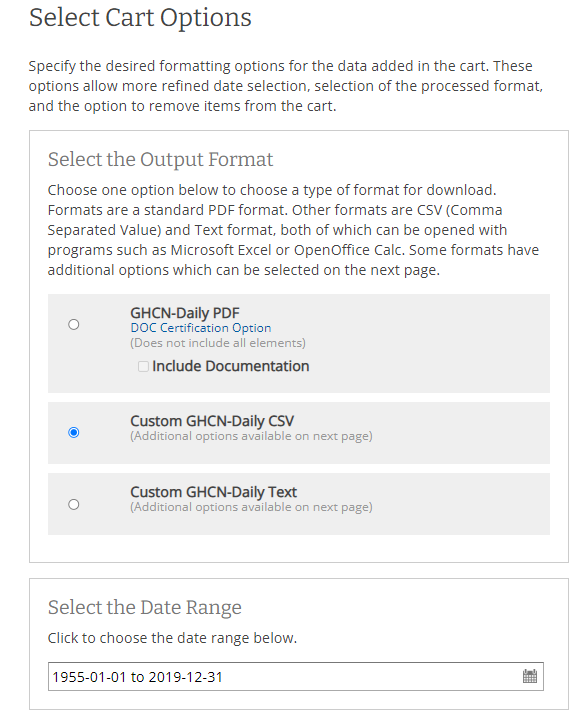
- At the custom options we select the fields we want to have in our dataset and the units of our data
(metric). For our case we should only select data about temperature and precipitation, but I select
all the fields just to be safe
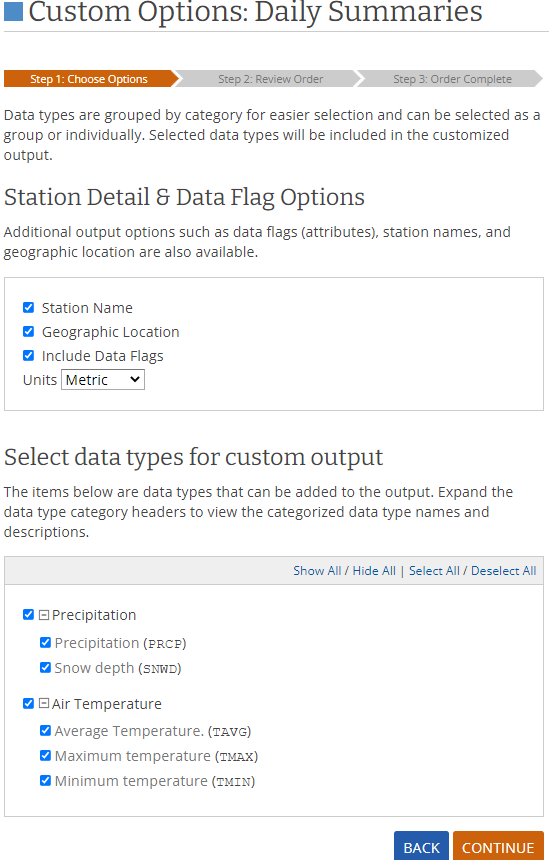
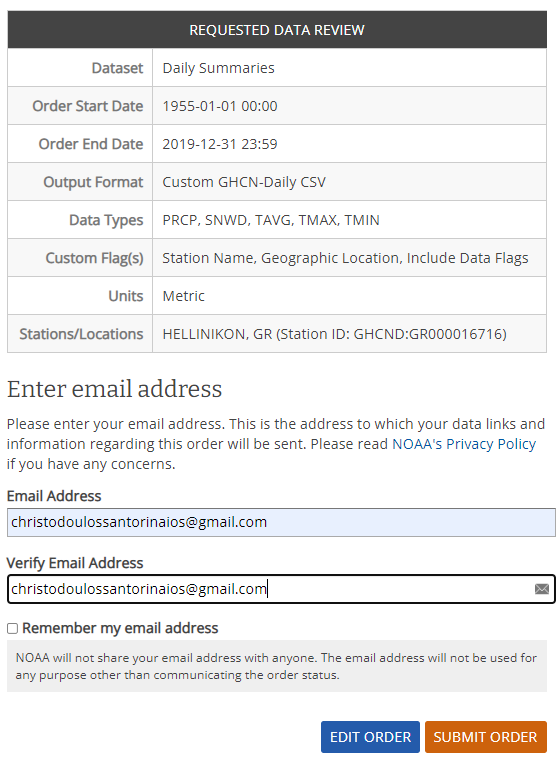
- In the page that loads we can confirm our selections and place our order by inputting a valid email
address. It's safe to input a personal or business email address here as NOAA will use the email
address for the sole purpose of communicating the order status to us.
- After submitting the request we are informed that the order is complete and get various information
about the dataset. After a couple of minutes we receive a link to our email with the dataset or we
can check order status and the download link will be there.
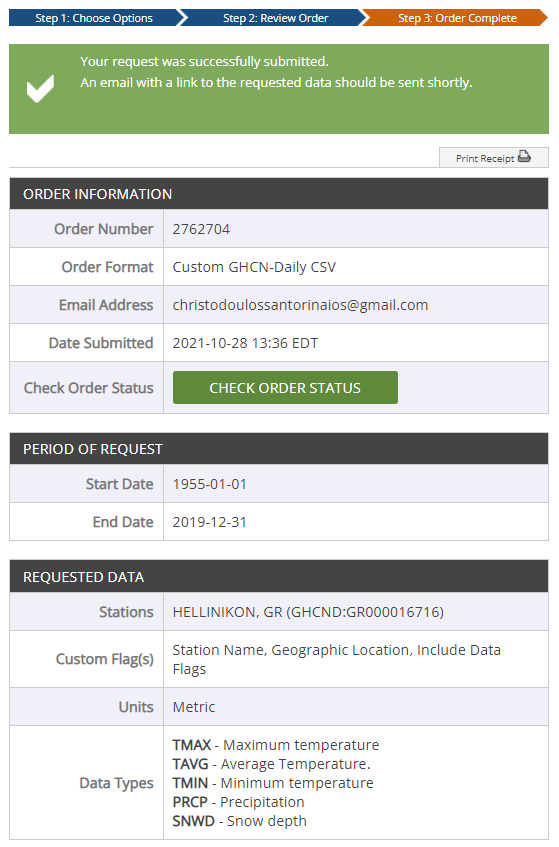
We have two options here. We can use the download link to connect to the csv online (although it expires after 30 days) or we can download and work with dataset locally. I chose the second option.
Now that we have the data, let's import the libraries we are going to use in our analysis.
#import required libraries
%matplotlib inline
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib
import seaborn as sns
matplotlib.rcParams['figure.dpi'] = 300
We read the data and it's important here to use the parse_dates argument to get the dates as datetime64 objects.
# read csv as dataframe parsing dates
df = pd.read_csv('noaa.csv', parse_dates=['DATE'])
Let's have a look on our data, descriptive statistics, missing values, etc. to get a first understanding of what's going on.
df.shape
(22885, 16)
df.sample(10)
| STATION | NAME | LATITUDE | LONGITUDE | ELEVATION | DATE | PRCP | PRCP_ATTRIBUTES | SNWD | SNWD_ATTRIBUTES | TAVG | TAVG_ATTRIBUTES | TMAX | TMAX_ATTRIBUTES | TMIN | TMIN_ATTRIBUTES | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8359 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1977-11-20 | 15.9 | ,,E | NaN | NaN | 15.9 | H,,S | 18.6 | ,,E | 12.1 | ,,E |
| 2476 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1961-10-12 | 0.0 | ,,E | NaN | NaN | 16.1 | H,,S | 18.6 | ,,E | 14.3 | ,,E |
| 17224 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 2002-02-27 | 0.0 | ,,E | NaN | NaN | 12.9 | H,,S | 17.0 | ,,E | 9.2 | ,,E |
| 8178 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1977-05-23 | 0.0 | ,,E | NaN | NaN | 23.9 | H,,S | 27.6 | ,,E | 19.8 | ,,E |
| 3632 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1964-12-11 | 0.0 | ,,E | NaN | NaN | 11.8 | H,,S | 12.0 | ,,E | 10.6 | ,,E |
| 9457 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1980-11-22 | 0.0 | ,,E | NaN | NaN | 10.7 | H,,S | 16.0 | ,,E | 5.4 | ,,E |
| 21512 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 2016-03-05 | 0.8 | ,,S | NaN | NaN | 13.3 | H,,S | 16.8 | ,,S | 8.6 | ,,S |
| 3238 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1963-11-13 | 0.0 | ,,E | NaN | NaN | 19.9 | H,,S | 23.6 | ,,E | 16.2 | ,,E |
| 14962 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1995-12-19 | 4.0 | ,,E | NaN | NaN | 13.6 | H,,S | 15.6 | ,,E | 9.2 | ,,E |
| 3785 | GR000016716 | HELLINIKON, GR | 37.9 | 23.75 | 10.0 | 1965-05-13 | 0.1 | ,,E | NaN | NaN | 15.4 | H,,S | 19.2 | ,,E | 13.0 | ,,E |
df.dtypes
STATION object NAME object LATITUDE float64 LONGITUDE float64 ELEVATION float64 DATE datetime64[ns] PRCP float64 PRCP_ATTRIBUTES object SNWD float64 SNWD_ATTRIBUTES object TAVG float64 TAVG_ATTRIBUTES object TMAX float64 TMAX_ATTRIBUTES object TMIN float64 TMIN_ATTRIBUTES object dtype: object
df.isna().sum()
STATION 0 NAME 0 LATITUDE 0 LONGITUDE 0 ELEVATION 0 DATE 0 PRCP 450 PRCP_ATTRIBUTES 450 SNWD 22856 SNWD_ATTRIBUTES 22856 TAVG 2311 TAVG_ATTRIBUTES 2311 TMAX 903 TMAX_ATTRIBUTES 903 TMIN 787 TMIN_ATTRIBUTES 787 dtype: int64
df.describe()
| LATITUDE | LONGITUDE | ELEVATION | PRCP | SNWD | TAVG | TMAX | TMIN | |
|---|---|---|---|---|---|---|---|---|
| count | 2.288500e+04 | 22885.00 | 22885.0 | 22435.000000 | 29.000000 | 20574.000000 | 21982.000000 | 22098.000000 |
| mean | 3.790000e+01 | 23.75 | 10.0 | 1.017174 | 157.379310 | 18.255366 | 22.325457 | 14.456535 |
| std | 7.105583e-15 | 0.00 | 0.0 | 4.658466 | 359.709547 | 6.927408 | 7.389128 | 6.464360 |
| min | 3.790000e+01 | 23.75 | 10.0 | 0.000000 | 10.000000 | -2.000000 | 1.000000 | -4.200000 |
| 25% | 3.790000e+01 | 23.75 | 10.0 | 0.000000 | 10.000000 | 12.700000 | 16.500000 | 9.400000 |
| 50% | 3.790000e+01 | 23.75 | 10.0 | 0.000000 | 20.000000 | 17.700000 | 21.800000 | 14.200000 |
| 75% | 3.790000e+01 | 23.75 | 10.0 | 0.000000 | 41.000000 | 24.300000 | 28.700000 | 20.000000 |
| max | 3.790000e+01 | 23.75 | 10.0 | 142.000000 | 1240.000000 | 34.800000 | 42.000000 | 30.400000 |
Some first remarks about our data:
- The types of the columns are in the format that we are ok to work with
- There are 450 missing values in precipitation and 2311 in TAVG. These are values we should handle as they are needed for our analysis
- There missing values in TMAX and TMIN as well. We will try ti handle them as well because they might be useful to address the missing average temperature values
- Almost all the values about snowfall are missing but it's something that doesn't concern us because we won't using them in our analysis
As a matter of fact let's go on and create a new dataframe with the columns that are going to be used later on. We don't need any of the station information of course. This dataframe will be a copy of the columns we need from the first one.
df_TEMP_PRCP = df[['DATE', 'TAVG', 'TMIN', 'TMAX', 'PRCP']].copy()
df_TEMP_PRCP.sample(10)
| DATE | TAVG | TMIN | TMAX | PRCP | |
|---|---|---|---|---|---|
| 6100 | 1971-09-14 | NaN | 20.3 | 28.8 | 0.0 |
| 20089 | 2010-01-04 | 8.9 | 5.8 | 11.0 | 0.0 |
| 16610 | 2000-06-23 | 26.3 | 19.0 | 31.8 | 0.0 |
| 9575 | 1981-03-20 | 13.4 | 11.0 | 18.4 | 0.0 |
| 14384 | 1994-05-20 | 21.2 | 18.0 | 26.0 | 0.0 |
| 15798 | 1998-04-03 | 14.3 | 10.0 | 20.0 | 0.0 |
| 18153 | 2004-09-13 | 21.1 | 13.0 | 27.2 | 0.0 |
| 10646 | 1984-02-24 | 12.0 | 8.4 | 16.0 | 0.0 |
| 18741 | 2006-04-26 | 17.3 | 12.4 | 22.2 | 0.0 |
| 16221 | 1999-05-31 | 23.2 | 18.6 | 28.8 | 0.0 |
df_TEMP_PRCP.isna().sum()
DATE 0 TAVG 2311 TMIN 787 TMAX 903 PRCP 450 dtype: int64
We will now try to fill the missing values on our dataset to the best of our ability. For this purpose, let's download another dataset that will help us with information about 2010-2019. This is the dataset from the Hellenic Data Service found here. We are interested in athens.csv as we want to use it to fill data of Hellinikon Station located in Athens.
# load athens.csv to dataframe, header=None because with it it reads the data in the first row as headers
df_hds = pd.read_csv('https://data.hellenicdataservice.gr/dataset/d3b0d446-aaba-49a8-acce-e7c6f6f5d3b5/resource/a7c024b3-8606-4f08-93e2-2042f5bd6748/download/athens.csv', header=None, parse_dates=[0])
df_hds.shape
(3652, 14)
df_hds.sample(10)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2158 | 2015-11-29 | 14.9 | 15.0 | 14.8 | 68.3 | 79 | 55 | 1018.9 | 1021.6 | 1016.7 | 0.0 | 1.4 | SW | 5.8 |
| 1424 | 2013-11-25 | 15.0 | 15.1 | 14.9 | 74.2 | 90 | 53 | 1007.8 | 1008.7 | 1007.3 | 0.2 | 0.7 | N | 3.4 |
| 1671 | 2014-07-30 | 29.6 | 29.7 | 29.5 | 60.6 | 71 | 47 | 1010.2 | 1011.1 | 1009.0 | 0.0 | 2.6 | SW | 8.1 |
| 1737 | 2014-10-04 | 19.0 | 19.0 | 18.9 | 58.4 | 66 | 52 | 1019.7 | 1020.5 | 1018.5 | 0.0 | 9.0 | NNE | 18.7 |
| 936 | 2012-07-25 | 30.1 | 30.3 | 30.0 | 56.1 | 71 | 42 | 1009.5 | 1010.9 | 1008.2 | 0.0 | 3.2 | SSW | 9.7 |
| 182 | 2010-07-02 | 28.0 | 28.2 | 27.9 | 53.7 | 67 | 39 | 1012.3 | 1013.1 | 1011.1 | 0.0 | 3.0 | NNW | 8.9 |
| 1377 | 2013-10-09 | 19.7 | 19.8 | 19.6 | 60.7 | 78 | 35 | 1023.4 | 1025.5 | 1021.7 | 0.0 | 1.8 | SSW | 6.3 |
| 3007 | 2018-03-27 | 15.6 | 15.7 | 15.5 | 62.1 | 78 | 49 | 1012.4 | 1013.7 | 1011.4 | 0.0 | 2.3 | WSW | 7.7 |
| 3508 | 2019-08-10 | 31.7 | 31.8 | 31.6 | 37.4 | 46 | 31 | 1010.8 | 1011.7 | 1009.5 | 0.0 | 11.3 | NNE | 25.6 |
| 849 | 2012-04-29 | 22.2 | 22.3 | 22.0 | 47.6 | 73 | 31 | 1015.3 | 1016.5 | 1013.4 | 0.0 | 3.1 | SW | 9.1 |
We immediately notice that the column headers don't have labels. So, we need to find some documentation about this. Fortunately, here the dataset is described.
Each line consists of 14 columns. First is the date of the parameters (YYYY-MM-DD). Next are the mean, maximum and minimum temperature (°C). After that and in the same order we have 6 columns for relative humidity and atmospheric pressure (%, hPa). The tenth column is the daily rainfall (mm). In the last 3 columns we can see mean wind speed, dominant wind direction as well as wind gust (km/h).
Let's now rename the columns.
df_hds.rename(columns={0: 'DATE', 1: 'TAVG', 2: 'TMAX', 3: 'TMIN', 4: 'HUMAVG', 5: 'HUMMAX', 6: 'HUMMIN', 7: 'ATMAVG',
8: 'ATMMAX', 9: 'ATMMIN', 10: 'RAIN', 11: 'WINDAVG', 12: 'WINDDIR', 13: 'WINDGUST'}, inplace=True)
df_hds.head()
| DATE | TAVG | TMAX | TMIN | HUMAVG | HUMMAX | HUMMIN | ATMAVG | ATMMAX | ATMMIN | RAIN | WINDAVG | WINDDIR | WINDGUST | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2010-01-01 | 17.9 | 18.1 | 17.8 | 61.4 | 91 | 33 | 1003.6 | 1006.3 | 1002.0 | 0.2 | 4.0 | WSW | 12.7 |
| 1 | 2010-01-02 | 15.6 | 15.7 | 15.5 | 57.4 | 70 | 45 | 1005.2 | 1008.7 | 1001.5 | 0.0 | 6.8 | WSW | 20.7 |
| 2 | 2010-01-03 | 13.5 | 13.6 | 13.4 | 56.0 | 76 | 39 | 1011.7 | 1016.7 | 1008.6 | 0.0 | 5.0 | WSW | 15.4 |
| 3 | 2010-01-04 | 9.5 | 9.6 | 9.5 | 50.7 | 60 | 38 | 1021.3 | 1023.1 | 1016.8 | 0.0 | 4.3 | NNE | 11.0 |
| 4 | 2010-01-05 | 13.4 | 13.5 | 13.4 | 70.5 | 82 | 54 | 1018.7 | 1022.1 | 1015.5 | 0.0 | 7.9 | S | 19.8 |
Let's keep the columns we are interested in by taking a copy of this dataframe. Note that precipitation is the rain variable in the second dataset so let's change the column name.
df_hds.rename(columns={'RAIN': 'PRCP'}, inplace=True)
df_hds_TEMP_PRCP = df_hds[['DATE', 'TAVG', 'TMAX', 'TMIN', 'PRCP']].copy()
df_hds_TEMP_PRCP.isna().sum()
DATE 0 TAVG 0 TMAX 0 TMIN 0 PRCP 0 dtype: int64
There are no missing values in the second dataset. Now, let's look what we can cover from our original dataset.
There are no missing values in average temperature but there are missing values in max temperature, min temperature and precipitation in the first dataset.
Since we are going to combine data from different dataframes and we are working with dates, a good idea would be to turn the DATES column to index column in both dataframes.
df_TEMP_PRCP.set_index(['DATE'], inplace=True)
df_hds_TEMP_PRCP.set_index(['DATE'], inplace=True)
While trying to find the average I got this error:
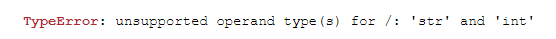 I didn't expect to get an error here and I started searching for the issue as I thought that everything
I had was numbers.
Running df_TEMP_PRCP.dtypes got me:
I didn't expect to get an error here and I started searching for the issue as I thought that everything
I had was numbers.
Running df_TEMP_PRCP.dtypes got me:
 I had seen that I had only float64 objects in df_TEMP_PRCP at the start so the problem should have been
in df_hds_TEMP_PRCP. Indeed, there the temperatures were objects and not float64 and when I tried to
convert them got this error message:
I had seen that I had only float64 objects in df_TEMP_PRCP at the start so the problem should have been
in df_hds_TEMP_PRCP. Indeed, there the temperatures were objects and not float64 and when I tried to
convert them got this error message:
 Ok, that was unexpected. There were some dashes in the data in some columns and I didn't find them as
they are not considered na.
I located these rows.
Ok, that was unexpected. There were some dashes in the data in some columns and I didn't find them as
they are not considered na.
I located these rows.
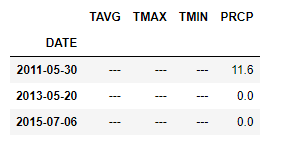 Then checked in the first dataset, if I have any values for them.
Then checked in the first dataset, if I have any values for them.
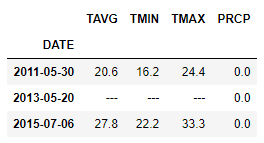 I had in the two of the three rows, so here I choose to drop them and move on with my analysis.
I had in the two of the three rows, so here I choose to drop them and move on with my analysis.
df_hds_TEMP_PRCP[(df_hds_TEMP_PRCP.TMIN == '---') | (df_hds_TEMP_PRCP.TMAX == '---') | (df_hds_TEMP_PRCP.TAVG == '---')].index
DatetimeIndex(['2011-05-30', '2013-05-20', '2015-07-06'], dtype='datetime64[ns]', name='DATE', freq=None)
convert_dict = {'TMIN': float,
'TMAX': float,
'TAVG': float
}
df_hds_TEMP_PRCP.drop(labels=[pd.Timestamp('2011-05-30 00:00:00'), pd.Timestamp('2013-05-20 00:00:00'), pd.Timestamp('2015-07-06 00:00:00')], inplace=True)
df_hds_TEMP_PRCP = df_hds_TEMP_PRCP.astype(convert_dict)
df_hds_TEMP_PRCP.dtypes
TAVG float64 TMAX float64 TMIN float64 PRCP float64 dtype: object
We now fill the na values of the df_TEMP_PRCP dataframe with the values from the df_hds_TEMP_PRCP dataframe.
df_TEMP_PRCP.fillna(df_hds_TEMP_PRCP, inplace=True)
df_TEMP_PRCP[(df_TEMP_PRCP.index.year>=2010) & (df_TEMP_PRCP.index.year<2020)]
| TAVG | TMIN | TMAX | PRCP | |
|---|---|---|---|---|
| DATE | ||||
| 2010-01-01 | 18.0 | 17.8 | 21.8 | 0.2 |
| 2010-01-02 | 16.0 | 13.8 | 17.8 | 0.0 |
| 2010-01-03 | 13.5 | 10.8 | 15.6 | 0.0 |
| 2010-01-04 | 8.9 | 5.8 | 11.0 | 0.0 |
| 2010-01-05 | 13.1 | 8.6 | 16.0 | 0.0 |
| ... | ... | ... | ... | ... |
| 2019-12-27 | 9.8 | 6.2 | 13.4 | 0.0 |
| 2019-12-28 | 8.6 | 8.2 | 10.8 | 0.0 |
| 2019-12-29 | 5.9 | 3.8 | 8.0 | 17.5 |
| 2019-12-30 | 4.2 | 3.9 | 7.7 | 4.6 |
| 2019-12-31 | 5.9 | 2.8 | 6.5 | 37.6 |
2799 rows × 4 columns
Perfect, now the data in the columns of df_TEMP_PRCP are filled and we have everything we need... Well, that is almost the case. Notice though that there 2799 rows in the dataframe instead of 3649 that should have been there (noticed this in the number of rows of the df_hds dataframe). So, let's go on and fill these rows as well. To do this, we find the difference in indexes between the two dataframes and then we concatenate them.
index_difference = df_hds_TEMP_PRCP.index.difference(df_TEMP_PRCP.index)
len(index_difference)
852
The length of the difference is 852 as expected but it's nice when we take the time to confirm things.
df_TEMP_PRCP = pd.concat([df_TEMP_PRCP, df_hds_TEMP_PRCP.loc[index_difference]], axis=0).sort_index()
df_TEMP_PRCP[(df_TEMP_PRCP.index.year>=2010) & (df_TEMP_PRCP.index.year<2020)].shape
(3651, 4)
df_TEMP_PRCP[(df_TEMP_PRCP.index.year>=2010) & (df_TEMP_PRCP.index.year<2020)].isna().sum()
TAVG 0 TMIN 0 TMAX 0 PRCP 0 dtype: int64
For now, it seems ok and we don't need the second dataframe anymore.
As a sidenote, I noticed that there were discrepancies in the values of the second dataset to the values of the first. For our analysis, I choose to keep the values of the first set and only fill missing values from the second. I could take their average or keep the values from the second dataset, but I choose not to as the purpose of our initial research was only to fill missing values.
Let's check the dataframe now.
df_TEMP_PRCP.isna().sum()
TAVG 2311 TMIN 338 TMAX 83 PRCP 348 dtype: int64
There are still missing values. We will try to cover the TAVG by taking the average of TMIN and TMAX. This, of course, is not totally correct as in a day we could have values near TMIN or TMAX for most of the day so the average isn't accurate but just to have an estimation and better fill our data.
df_TEMP_PRCP.TAVG = df_TEMP_PRCP.TAVG.fillna((df_TEMP_PRCP.TMIN + df_TEMP_PRCP.TMAX)/2)
df_TEMP_PRCP.dtypes
TAVG float64 TMIN float64 TMAX float64 PRCP float64 dtype: object
df_TEMP_PRCP.isna().sum()
TAVG 1 TMIN 338 TMAX 83 PRCP 348 dtype: int64
df_TEMP_PRCP[df_TEMP_PRCP.TAVG.isna()]
| TAVG | TMIN | TMAX | PRCP | |
|---|---|---|---|---|
| DATE | ||||
| 1957-03-09 | NaN | NaN | 16.9 | 0.3 |
Q2: Deviation of Summer Temperatures¶
The Hellenic National Meteorological Service has published a report on extreme weather events for 2020. The report is available at http://www.hnms.gr/emy/en/pdf/2020_GRsignificantEVENT_en.pdf. In page 7 of the report there is a graph showing the mean summer temperature deviation from a baseline of 1971-2000.
We will try to create our own version of the graph, using a baseline of 1974-1999. The graph should look like the one below. The line that runs through the graph is the 10 years rolling average of the deviation from the mean. How can we intepret this figure?
We need to calculate the baseline mean temperature for the summers of 1974-1999. So, let's get the mean summer temperatures of these years.
baseline_mean_temp = df_TEMP_PRCP[(df_TEMP_PRCP.index.year>=1974) & (df_TEMP_PRCP.index.year<=1999) & (df_TEMP_PRCP.index.month>=6) & (df_TEMP_PRCP.index.month<=8)].TAVG.mean()
df_TEMP_PRCP_summer = df_TEMP_PRCP[(df_TEMP_PRCP.index.month>=6) & (df_TEMP_PRCP.index.month<=8)]
df_summer_means = df_TEMP_PRCP_summer['TAVG'].groupby(df_TEMP_PRCP_summer.index.year).mean()
df_summer_means_dif = df_summer_means - baseline_mean_temp
df_summer_means_dif
DATE
1955 -0.279640
1956 0.912751
1957 1.141012
1958 0.850794
1959 0.170360
...
2015 1.092099
2016 1.923620
2017 1.857316
2018 1.206229
2019 1.804055
Name: TAVG, Length: 65, dtype: float64
df_rolling = df_summer_means_dif.rolling(window=10, min_periods=1).mean()
from IPython.core.display import HTML
HTML("""
<style>
.output_png {
display: table-cell;
text-align: center;
vertical-align: middle;
}
</style>
""")
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,12))
ax.set_axisbelow(True)
plt.rcParams.update({'axes.titlesize': 'x-large'})
ax.set_title('Mean Summer Temperature Difference from the 1974-1999 Mean', {'fontsize': plt.rcParams['axes.titlesize']})
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
colors = ['#FF8C00' if i>0 else "#1111FD" for i in df_summer_means_dif]
plt.bar(df_summer_means_dif.index, df_summer_means_dif, width=0.2, color=colors)
ax.set_facecolor('#E5E5E5')
plt.grid(color='w')
plt.plot([1955, 2019], [0, 0], color='#777777')
plt.plot(df_rolling.index, df_rolling, color='#B5A3DB')
[<matplotlib.lines.Line2D at 0x27d99b8e500>]
The figure shows us that years from 1956 to 1968 we hotter than the average, then there were two and a half decades of colder than the average summers and in the last almost 30 years there is a steady rise of the mean summers temperature, something that is really alarming and concides with the warnings scientists give about global warming.
Q3: Evolution of Daily Temperatures¶
Now, we will get the average temperate for each year for the full period from 1955 to 2020. We then create a plot showing the daily temperature for each year. The line corresponding to each year will be smoothed by using a 30 days rolling average. The lines are colored from light orange to dark orange, progressing through the years in ascending order.
On that plot we overlay a line showing the average daily temperature for the baseline period of 1974-1999 (that is the black line). The line will also be smoothed usng a 30 years rolling average. How can we interpret this figure?
df_TEMP_PRCP.index.date
array([datetime.date(1955, 1, 1), datetime.date(1955, 1, 2),
datetime.date(1955, 1, 3), ..., datetime.date(2019, 12, 29),
datetime.date(2019, 12, 30), datetime.date(2019, 12, 31)],
dtype=object)
Here we set the index as year and day of year and then unstack the series object in order to keep only year as index and have the days as columns.
df_TEMP = df_TEMP_PRCP.set_index([df_TEMP_PRCP.index.year, df_TEMP_PRCP.index.dayofyear]).TAVG
df_TEMP.index.rename(['year', 'dayOfYear'], inplace=True)
df_TEMP = df_TEMP.unstack()
df_TEMP
| dayOfYear | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 357 | 358 | 359 | 360 | 361 | 362 | 363 | 364 | 365 | 366 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| year | |||||||||||||||||||||
| 1955 | 14.35 | 10.70 | 12.70 | 13.05 | 13.15 | 14.25 | 13.30 | 15.80 | 14.7 | 13.2 | ... | 14.85 | 14.25 | 12.0 | 11.60 | 10.2 | 8.55 | 8.9 | 9.25 | 13.80 | NaN |
| 1956 | 16.35 | 14.50 | 12.15 | 11.65 | 8.60 | 7.15 | 9.15 | 10.35 | 10.3 | 8.7 | ... | 11.00 | 9.80 | 14.3 | 12.40 | 10.3 | 11.20 | 8.3 | 9.00 | 9.15 | 8.15 |
| 1957 | 9.80 | 10.30 | 10.40 | 8.10 | 8.90 | 8.55 | 9.65 | 8.60 | 7.4 | 9.5 | ... | 8.90 | 9.90 | 9.3 | 10.20 | 10.2 | 9.80 | 11.9 | 15.70 | 15.80 | NaN |
| 1958 | 12.10 | 11.80 | 12.10 | 10.30 | 8.50 | 10.20 | 13.60 | 15.60 | 7.8 | 5.8 | ... | 16.80 | 16.40 | 15.3 | 12.90 | 13.6 | 12.40 | 13.5 | 11.20 | 9.00 | NaN |
| 1959 | 10.90 | 12.45 | 13.70 | 15.50 | 12.55 | 8.20 | 7.15 | 9.40 | 13.5 | 12.5 | ... | 12.30 | 11.85 | 13.0 | 12.15 | 11.8 | 13.55 | 13.6 | 10.50 | 10.35 | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 2015 | 4.40 | 4.50 | 5.50 | 11.10 | 6.20 | 3.30 | 3.40 | 4.20 | 6.8 | 13.3 | ... | 13.00 | 12.60 | 13.0 | 12.50 | 12.4 | 11.80 | 12.4 | 9.90 | 3.40 | NaN |
| 2016 | 3.80 | 7.90 | 11.50 | 13.40 | 17.20 | 16.40 | 14.10 | 13.40 | 13.2 | 13.2 | ... | 6.80 | 7.90 | 9.2 | 9.10 | 11.2 | 10.80 | 7.2 | 3.90 | 3.60 | 3.40 |
| 2017 | 6.00 | 8.40 | 11.10 | 11.80 | 13.20 | 10.60 | 2.30 | 0.50 | 0.8 | 2.5 | ... | 6.50 | 7.90 | 10.6 | 11.30 | 12.9 | 14.30 | 12.7 | 11.30 | 9.70 | NaN |
| 2018 | 11.30 | 13.70 | 11.50 | 9.60 | 10.40 | 11.60 | 13.90 | 15.20 | 14.0 | 13.2 | ... | 13.50 | 12.50 | 9.6 | 6.00 | 6.9 | 8.80 | 10.8 | 10.60 | 9.70 | NaN |
| 2019 | 8.30 | 7.40 | 6.40 | 5.70 | 3.90 | 4.80 | 4.40 | 2.20 | 8.6 | 13.3 | ... | 14.50 | 12.70 | 11.3 | 10.60 | 9.8 | 8.60 | 5.9 | 4.20 | 5.90 | NaN |
65 rows × 366 columns
Next, we calculate the rolling average with a window of 30 days.
df_TEMP_rolling = df_TEMP.rolling(window=30).mean()
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(16,6))
ax.set_axisbelow(True)
ax.set_ylabel('Aberage Daily Temperature')
ax.set_xlabel('Date')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.set_facecolor('#E5E5E5')
plt.grid(color='w')
# got this code snippet from StackOverflow. It maps the days of the year to the appropriate months
import datetime as dt
import matplotlib.dates as mdates
start = dt.datetime(2016,1,1) #there has to be a year given, even if it isn't plotted
new_dates = [start + dt.timedelta(days=i) for i in range(366)]
x = new_dates
y = df_TEMP.index
xfmt = mdates.DateFormatter('%b')
months = mdates.MonthLocator()
ax.xaxis.set_major_locator(months)
ax.xaxis.set_major_formatter(xfmt)
# Now we plot our lines usings a loop to get the rows one by one
for _, year in df_TEMP_rolling.iterrows():
plt.plot(year.index, year.values)
The interpretation of the figure is that the weather is hotter in the summer months and we also notice that darker orange lines are higher. That means that the recent years have been hotter on average than past years.
Q4: Extreme Temperature Events¶
Another measure used by climatologists is the number of extreme events. Extreme events are defined as those beyond 5% or 10% from the expected value. We will deal with extreme heat events going 10% above the baseline.
We will count the number of extreme temperature events per year, compared to the baseline of 1974-1999. We should be able to produce a graph like the one that follows. The vertical axis is the percentage of extreme heat events calculated over the number of observations for each year. The gray line in the middle is the average percentage of extreme tempearture events of the baseline. The colour blue is used for those years where the percentage is below the baseline; otherwise the colour is orange. How can we interpret this graph?
We calculate the baseline of the temperatures from 1974-1999.
baseline = df_TEMP_PRCP[(df_TEMP_PRCP.index.year>=1974) & (df_TEMP_PRCP.index.year<=1999)].TAVG.mean()
baseline
17.852632687447347
Here we get the days with temperatures above the 110% of the baseline and then we count them.
df_extreme = df_TEMP_PRCP[df_TEMP_PRCP.TAVG > 1.1 * baseline]
df_extreme_days = df_extreme.groupby(df_extreme.index.year).count()
df_extreme_days['years'] = df_extreme_days.index
Calculate number of extreme days between 1974 and 1999.
number_of_extreme_days_74_99 = df_extreme_days[(df_extreme_days.index>=1974) & (df_extreme_days.index<=1999)].TAVG.sum()
# importing calendat to get leap years
import calendar
df_extreme_days['number_of_days_in_year'] = df_extreme_days.years.apply(lambda x: 366 if calendar.isleap(x) else 365)
df_extreme_days['percentage'] = df_extreme_days.TAVG / df_extreme_days.number_of_days_in_year
number_of_baseline_days = df_extreme_days[(df_extreme_days.index>=1974) & (df_extreme_days.index<=1999)]['number_of_days_in_year'].sum()
Calculate baseline percentage.
baseline_percentage = number_of_extreme_days_74_99 / number_of_baseline_days
baseline_percentage
0.413647851727043
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,10))
ax.set_axisbelow(True)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.set_facecolor('#E5E5E5')
plt.grid(color='w')
colors = ['#FF8C00' if i>baseline_percentage else "b" for i in df_extreme_days.percentage]
plt.bar(df_extreme_days.index, df_extreme_days.percentage, width=0.2,color=colors)
plt.axhline(y=baseline_percentage, color='#777777')
<matplotlib.lines.Line2D at 0x23feb68aef0>
The interpretation of the graph is that in recent years extreme heat events are much more prevalent than in the past.
Q5: Precipitation¶
Continuing the thread on extreme events, another consideration is rainfall. The weather may or may not be drying up. We are, however, interested in whether precipication becomes more intense over time.
To see that, we will count the overall rainfall over the year and the number of rainy days in each year. Then, by dividing the rainfall by the number of rainy days we will get an indication of whether we are getting rain in more concentrated bursts. We will then create a plot showing the ratio of rainfall over rainy days over the years. On the plot we will overlay the 10 years rolling average. The plot should be similar to the one below. How can we interpret this plot?
Firstly, we get the sum of the precipitation for each year.
prcp_sum = df_TEMP_PRCP.groupby(df_TEMP_PRCP.index.year).sum()['PRCP']
Then we make a dataframe with only the days that had some rainfall and count how mnay those days were per year.
df_PRCP = df_TEMP_PRCP[df_TEMP_PRCP.PRCP > 0]
df_PRCP.groupby(df_PRCP.index.year).count()
| TAVG | TMIN | TMAX | PRCP | |
|---|---|---|---|---|
| DATE | ||||
| 1955 | 68 | 68 | 68 | 68 |
| 1956 | 52 | 52 | 52 | 52 |
| 1957 | 72 | 72 | 73 | 73 |
| 1958 | 68 | 68 | 68 | 68 |
| 1959 | 62 | 62 | 62 | 62 |
| ... | ... | ... | ... | ... |
| 2015 | 41 | 41 | 41 | 41 |
| 2016 | 42 | 42 | 42 | 42 |
| 2017 | 48 | 48 | 48 | 48 |
| 2018 | 55 | 55 | 55 | 55 |
| 2019 | 58 | 58 | 58 | 58 |
65 rows × 4 columns
prcp_num_days = df_TEMP_PRCP[df_TEMP_PRCP.PRCP > 0].groupby(df_TEMP_PRCP[df_TEMP_PRCP.PRCP > 0].index.year).count()['PRCP']
We divide the sum of the rainfall amount in mm of every year with the number that it rained in every year.
prcp_burst = prcp_sum / prcp_num_days
prcp_burst.index
Int64Index([1955, 1956, 1957, 1958, 1959, 1960, 1961, 1962, 1963, 1964, 1965,
1966, 1967, 1968, 1969, 1970, 1971, 1972, 1973, 1974, 1975, 1976,
1977, 1978, 1979, 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987,
1988, 1989, 1990, 1991, 1992, 1993, 1994, 1995, 1996, 1997, 1998,
1999, 2000, 2001, 2002, 2003, 2004, 2005, 2006, 2007, 2008, 2009,
2010, 2011, 2012, 2013, 2014, 2015, 2016, 2017, 2018, 2019],
dtype='int64', name='DATE')
We calculate the rolling average.
prcp_rolling = prcp_burst.rolling(window=10, min_periods=1).mean()
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12,10))
ax.set_axisbelow(True)
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.set_facecolor('#E5E5E5')
plt.grid(color='w')
plt.bar(prcp_burst.index, prcp_burst,width=0.3, color='b')
plt.plot(prcp_rolling.index, prcp_rolling)
[<matplotlib.lines.Line2D at 0x23feeb00220>]
My interpretation of the graph is that there is some steady increase in the intensity of the rainfall as we go from the 60's to today.
Parting thoughts¶
I see that my graphs have a lot of differences to the given graphs. I believe this has to do with the hypothesis that I did early in the study that I can create many average temperatures by averaging min and max temperatures per day. Many of these temperatures created could be way off and play significant role in how the graphs are produced. Maybe it could be safe to leave them as NaN and just have some missing lines in the graphs as the ones depicted.



